Library Quant
Generation sequencing library quantification we recommend Q-PCR quantification.
Catalog # NQ101, NQ102, NQ103, NQ104, NQ105, NQ106
1. Introduction
VAHTS Library Quantification Kit
for Illumina®
VAHTS Library Quantification Kit is specially designed for accurate NGS quantification of the concentration of a DNA library in Illumina® next generation
sequencing (NGS) using SYBR Green qPCR. The kit includes DNA standard of known concentration that is used to generate a standard curve to which the
library samples are compared. VAHTS SYBR® qPCR Master Mix included in this kit is a new-type qPCR mix based on antibody modified hot-start, which is of
high specificity, high amplification efficiency, wide GC content adaptability, and high sensitivity. All kit components are subjected to stringent functional quality
control in order to guarantee the highest levels of stability and repeatability of library construction.
Note:
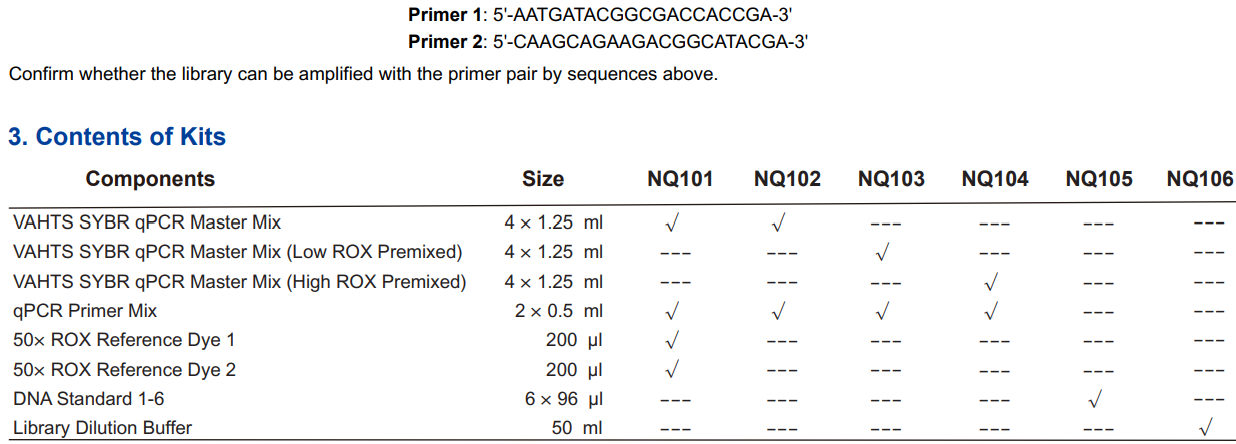
(1). NQ101, NQ102, NQ103, and NQ104 are amplification kits containing VAHTS SYBR qPCR Master Mix, qPCR Primer Mix, and individual ROX package
(only in NQ101). They are sufficient for 500 reactions (20 μl/reaction). NQ105 is a standard kit containing DNA Standard 1-6, which is sufficient for 8 standard
curves (3 repeated wells). NQ106 is Library Dilution Buffer (50 ml).
(2). The ROX Reference Dye is ssed to rectify the error of fluorescence signals between different wells. Select the appropriate ROX reference dye
according to the Real-time PCR instrument used:
Individual packages of both ROX Reference Dye 1 and 2 are provided in NQ101. Therefore, NQ101 is suitable for all quantitative PCR instruments.
Use
appropriate ROX Reference Dye according to the table above.
NQ102 kit contains no individual or pre-mixed ROX Reference Dye. It is only suitable for Type I qPCR instruments in the table above.
NQ103 kit contains Low ROX (ROX Reference Dye 2) in VAHTS SYBR qPCR Master Mix. It is suitable for Type III qPCR instruments in the table above.
NQ104 kit contains High ROX (ROX Reference Dye 1) in VAHTS SYBR qPCR Master Mix. It is suitable for Type qPCR instruments in the table above.
2. Applications
This kit is designed for the absolute quantification of the concentration of the Illumina® platform's NGS library. Regardless of the way you use for construction,
you can use this product for absolute quantification if the end of the library contains Illumina® P5 and P7 flow cell binding sequences. The library should be no
more than 1 kb in length and no less than 0.0002 pM in concentration. In addition, this product can also be used to detect the library pollution level in
experimental environment. Two kinds of primer sequences are provided in the qPCR Primer Mix in this kit:
Primer 1: 5'-AATGATACGGCGACCACCGA-3'
Primer 2: 5'-CAAGCAGAAGACGGCATACGA-3'
Confirm whether the library can be amplified with the primer pair by sequences above.
Version6.1
Vazyme Biotech Co., Ltd Order: global@vazyme.com Support: support@vazyme.com
www.vazyme.com For research use only, not for use in diagnostic procedures.
Components Size
4 × 1.25 ml
DO NOT USE ROX Reference Dye (Type I)
USE ROX Reference Dye 1 (Type II)
USE ROX Reference Dye 2 (Type III)
Bio-Rad CFX96™, CFX384™, iCycler iQ™, iQ™5, MyiQ™, MiniOpticon™, Opticon®
, Opticon 2, Chromo4™;
Cepheid SmartCycler®
; Eppendorf Mastercycler
® ep realplex, realplex 2 s; Illumina Eco qPCR;
Qiagen/Corbett Rotor-Gene
® Q, Rotor-Gene
® 3000, Rotor-Gene
® 6000;
Roche Applied Science LightCyclerTM 480; Thermo Scientific PikoReal Cycler.
Applied Biosystems 5700, 7000, 7300, 7700, 7900, 7900HT, 7900HT Fast; StepOne™, StepOnePlus™.
Applied Biosystems 7500, 7500 Fast, ViiA™7; Stratagene MX4000™, MX3005P™, MX3000P™.
All components should be stored at -20℃. The Master Mix and ROX should be protected from light.
The kits can be valid for 30 freeze-thaw cycles. Once be thawed, the Library Dilution Buffer and the Master Mix can be stored at 4℃ for up to 3 months.
4. Storage
Nucleoside Residue Detection: No activity of exonuclease or rendonuclease were detected in all components.
Standard Concentration Determination: Each batch of standards have been determined and checked by a variety of methods for repeated times. CV
values between batches are lower than 5%.
5. Quality Control
6. Notes
(1). Notes on Pipetting:
qPCR is extremely sensitive and is vulnerable to variation arising from a number of sources. Please read the following contents carefully before operation:
A. All components should be thawed and mixed thoroughly. Briefly centrifuge to collect liquid to the bottom before use.
B. DNA solution with high concentration is of very high viscosity and the DNA molecular dispersion is poor, please avoid directly dilution with a large volume
(such as 1:10000 dilution). Instead, serial dilutions with small volumes are recommended (i.e. dilute 1:100 twice to make a 1:10000 dilution).
C. Use filtered pipette tips to avoid aerosol contamination.
D. Do not use multichannel pipettors.
E. Change tips between samples to avoid cross contamination.
F. When pipetting, do not immerse the tip too deeply into the solution to avoid liquid sticking to the outside wall of the tip.
G. When pipetting the liquids out, keep the tips close to the bottom of the reaction tube.
H. Pipetting for 2-3 more times to rinse the tip after drawing out the solution.
I. After completely pipetting out the liquids, please check carefully to confirm there is no liquid remains in the tip.
(2). Library Concentration and Dilution Factor
The library must be diluted to the effective Ct range of standard curve for quantitative reaction. Ct values out of the effective range are not suitable for
quantification. If more than one dilution of each library is assayed, select Ct values within effective range for concentration calculation. Effective Ct range
selection scheme of standard curve can refer to Section 8. Data Analysis. The following table shows the concentration range of the quantitative library:
(3). Library Dilution
Dilute the DNA library with an appropriate buffer (Library Dilution Buffer, Vazyme, #NQ106; or self-provide buffer whith 10 mM Tris-HCl, pH 8.0 @25℃, 0.05%
Tween 20). DO NOT dilute the library with water. Use freshly diluted library for quantification. Before qPCR reaction, the diluted library and the thawed DNA
Standards should be kept on ice.
(4). Contaminations and No Template Controls (NTC)
A. Inappropriate operation may leads to contaminations in PCR products, which results in inaccurate quantitative results and low credibility. It is recommend to
physically isolate the sample preparation area from the template preparation area. Use specialized pipettor and filtered tips. Clean the experimental area
regularly (with 0.5% sodium hypochlorite or 10% bleaching agent).
B. Always dispense the DNA Standards from the lowest concentration to the highest (i.e. from DNA Standard 6 to DNA Standard 1). Replace tips after each
pipetting.
C. It is highly recommended that no-template controls (NTCs) are included in each assay to detect the PCR specificity and possible contamination introduced.
The primers used in this kit is based on universal sequences on Illumina® platform. It is normal that amplification product and a Ct value appears in NTC
reaction, due to inevitable aerosol pollutions during repeated library dilution. In this case, determine the effective CT range of the standard curve firstly
according to the NTC negative control (refer to Section 8. Data Analysis), and then draw the standard curve and calculate the concentration.
(5). Baseline Setting of the Amplification Curve
The concentration of DNA Standard 1 molar is significantly higher than that of the conventional qPCR templates, therefore generally, its Ct value is as small as
7-9 cycles. While for most qPCR instruments, the default baselines are set at 3-15 cycles, which may increase the Ct value of DNA Standard 1 by mistake and
may further affects the linear relationship of the standard curve. Manually set the baseline at 1-3 cycles to avoid this situation.
(6). Correction of Library Concentration
The fluorescence intensity of SYBR Green, a dye that binds to all double-stranded DNA molecules, is proportional to the molecular weight of DNA molecules.
For example, the fluorescence intensity of two 250 bp dsDNA molecules is equivalent to one 500 bp dsDNA molecule. Therefore, it is necessary to correct the
library length according to the length of DNA Standards (452 bp) and the average length of the library (refer to Section 8. Data Analysis).
(7). Melting Curve Analysis
It is recommended to collect melting curve data in every assay, because melt curve analysis is highly important to identify the maximum efficient Ct value and
contamination. The melt curves for the DNA Standards displays a characteristic double peak (as shown by the blue arrow). This is the result of differential local
melting in the 452 bp linear template and is not indicative of non-specific amplification. In addition, the molar concentrations of DNA Standards 1-3 are two
high, and the amplification products are too many to melt completely at the Tm. Therefore, it is normal that the melting curves of DNA Standards 1-3 sometimes
exhibit raising tails (as shown by the red arrow).
(8). Other Methods for Library Quantification
Methods for library quantification include approaches base on spectrophotometer (i.e. NanoDrop™), fluorescent dyes (i.e. Qubit®
, PicoGreen®
), electrophoresis
(i.e. 2100 Bioanalyzer, TapeStation, LabChip® GX), and qPCR. For amplified libraries with intact dual-index, the qPCR approach provides the most accurate
quantification results than other methods. Theoretically, library concentrations determined by qPCR are a slightly lower than that determined by other methods.
The reason is that qPCR measures only those molecules that can be amplified in PCR instead of total DNA. However, for over-amplified libraries, which
contains concentrations determined by qPCR are higher than other methods.
Molar Concentration:
Mass Concentration:
Copies Concentration:
20 - 0.0002 pM
5.5-0.000055pg/μl
12 × 106
- 12 × 101 copies/μl
7. Protocol
Vazyme Biotech Co., Ltd Order: global@vazyme.com Support: support@vazyme.com
www.vazyme.com For research use only, not for use in diagnostic procedures.
(1). Prepare an appropriate volume of library dilution buffer (refers to Section 6: Notes 3. Library Dilution).
Note: The Dilution Buffer is stored at 4℃. Equilibrate the dilution buffer to room temperature for 30 min before use and put it back to 4℃ after use.
(2). Dilute the library with Dilution Buffer. The optimum dilution ratio should be adjusted according to the library concentration. The recommended dilution ratio
is between 1:1,000 and 1:100,000. At least one additional 2-fold dilution of each library is also recommended (i.e. 1:10,000 and 1:20,000). Library should be
kept on ice and diluted freshly before each use.
(3). Prepare the reaction solution in a qPCR tube as follows:
*a. Only needed when using NQ101. Choose the appropriate ROX according to the qPCR instrument. Do not add it when using other kits and replace with H2O.
*b. Add H2O to the NTC negative control reaction tubes, add diluted library to the sample reaction tubes; and add DNA Standards into the standard curve reaction tubes. Always dispense
the DNA Standards from the lowest concentration to the highest (i.e. from DNA Standard 6 to DNA Standard 1) to avoid the aerosol pollution.
*c. The recommended reaction volume is 20 μl. For a 10 μl reaction system, please reduce the amount of each reagent in proportion.
(4). Run the following program for qPCR:
*a. If the average length of the library is above 600 bp, the annealing time should be extended from 45 sec to 90 sec.
*b. Program for melting curve may vary qPCR instruments. Please select the default melting curve program of the instrument used.
VAHTS SYBR qPCR Master Mix (Without ROX, Low or High ROX Premixed)
qPCR Primer Mix
ROX Reference Dye 1/2 *a
DNA Standard 1-6 *b
or Diluted Library
8. Data Analysis
(1). Standard Curve
A. Replicate data points should differ by ≤ 0.2 cycles. Filter the original Ct and calculate the average Ct value.
The Ct differentia between every two of the 3 repeats should be ≤ 0.2. If the Ct value of one repeat is significantly different from that of the other two others, this
Ct should be discard when calculate the average Ct value. If the differentia between every two of them was > 0.2, repeat the assay with particular focus on
improving pipetting accuracy.
B. Confirm the effective Ct range refer to the Ct of NTC.
If Ct (NTC) > Ct (DNA Standard 6) + 3, the Ct of DNA Standard 6 is the maximum effective CT. Generate the standard curve using the Ct values of DNA
Standard 1-6.
If Ct (DNA Standard 6) + 3 > Ct (NTC) > Ct (DNA Standard 5) + 3, the Ct of DNA Standard 5 is the maximum effective Ct. Generate the standard curve using
the Ct values of DNA Standard 1-5.
If Ct (DNA Standard 5) + 3 > Ct (NTC) > Ct (DNA Standard 4) + 3, the Ct of DNA Standard 4 is the maximum effective Ct. Generate the standard curve using
the Ct values of DNA Standard 1-4.
To guarantee the accuracy of quantification, please use at least four Ct values of DNA Standards to generate the standard curve. If Ct (DNA Standard 4) + 3 >
Ct (NTC), it’s indicated the serious contamination of the assay. It’s necessary to replace the all components of the assay, and repeat the tests.
C. Generate standard curves
Choose the Ct within the effective range (as ordinate) and the corresponding Log[pM] in the following table (as abscissa) to generate the standard curve. The
correlation coefficient R2 should not be less than 0.99, and the slope should be located between -3.1 and -3.6 (indicating the amplification efficiency lies
between 90% and 110%). If the standard curve parameters are poor, repeat the assay.
Standard Code
(2). Determination of Library Concentration
A. Replicate data points should differ by ≤ 0.2 cycles. Filter the original Ct and calculate the average Ct value.
The Ct differentia between every two of the replicates should ≤ 0.2. If the data set contains many outliers, results are unlikely to be reliable. Repeat the assay
with particular focus on improving pipetting accuracy.
03
Vazyme Biotech Co., Ltd Order: global@vazyme.com Support: support@vazyme.com
www.vazyme.com For research use only, not for use in diagnostic procedures.
B. Determination the concentration (pM) of the diluted library according to the standard curve.
Only Ct values of Dilution library Ct within the effective Ct range of standard curve can be used to calculate the concentration. Do not use the Ct beyond the
effective Ct range of standard curve to calculate the concentration of dilution library.
C. Correct the library length with the concentration of dilution Library (pM) according to the following formula.
Corrected concentration of diluted library (pM) = [452 bp/ average library length (BP)] × concentration of diluted library (pM)
D. Calculated the original library concentration (nM) according to the following formula.
Concentration of original library (nM) = Corrected concentration of diluted library (pM) × dilution factor / 1,000
Table 1. Ct Values of DNA Standards
9. Application Example
(1). Starting Materials
Two DNA libraries with insertion length of about 350 bp (the total length of the library is about 470 bp) are prepared using VAHTS Nano DNA Library Prep Kit
for Illumina®
(Vazyme, #ND601). The length and concentration of the library are detected by Agilent Bioanalyzer 2100 High Sensitivity DNA Assay (refers to
Table 2).
(2). Library Dilution
Both the two libraries are diluted in 1:10000 (serial dilution in 1:100 for twice) and 1:20000 (dilute the 1:10000 library in 1:2).
(3). Generate Standard Curve
A. The Ct difference between replicates should be ≤ 0.2. Filter the original Ct of DNA Standards. Remove the third data point of DNA Standard 6 (Table 1, "Ct"
column), and calculate the average Ct (Table 1, Average Ct" column).
B. Refer to the Ct of NTC, it can be inferred that the Ct of DNA Standard 6 is the maximum effective Ct. Generate a standard curve (Figure 1) by plotting the
average Ct values of DNA Standard 1-6 against molar concentrations (Table 1,"Log [pM]").
(4). Calculation of Library Concentration
A. The Ct difference between replicates should be ≤ 0.2. Filter the original Ct values of diluted library. Remove the second data point of Library 2, 1:20000
dilution (Table 2, "Library 2, 1:20000" column). Calculate the average Ct (Table 2, Row 5).
B. Calculate the concentration of diluted libraries (Table 2, Row 7) according to the standard curve.
C. Calculated corrected concentration of the diluted library according to the average length of the library (Table 2, Row 8).
D. Calculated original concentrations of diltued libraries (Table 2, Row 9) and the average original concentration of library (Table 2, Row 11) according to
d
10. Trouble Shooting
(1). The amplification efficiency is beyond the range of 90% - 110%
● The qPCR reaction system is contaminated if Ct (NTC) - Ct (DNA Standard 6) < 3 or Ct (DNA Standard 6) - Ct (DNA Standard 5) < 3.1, while the calculated
amplification efficiency is above 100%. Determine the source of contamination (Standard DNA or library DNA) by examining the melt curves of NTC.
● Inappropriate Baseline setting may delay Ct value of DNA Standard 1, further affecting the calculation of PCR efficiency. Manually adjust the baseline to 1-3
cycles.
● Poor accuracy in liquid handling.
(2). The R2 value of the standard curve is < 0.99.
● Poor accuracy in liquid handling.
● All reagents should be thawed and mix thoroughly before use.
● Ensure that the appropriate ROX reference dye was used.
(3). The amplification curve of standards are not evenly distributed.
● If Ct (DNA Standard 6) - Ct (DNA Standard 5) < 3.1, the reaction system is contaminated. Determine the source of contamination (Standard DNA or library DNA)
by examining the melt curves of NTC.
● If Ct (DNA Standard 2) - Ct (DNA Standard 1) < 3.1, the baseline setting inappropriate. Manually adjust the baseline to 1-3 cycles.
● If ΔCt among DNA Standards is > 3.6, the amplification efficiency is poor. Ensure that all reagents are thoroughly thawed and mixed before use. Confirm that
all reaction components were added at the correct concentration, and that the correct cycling protocol was used.
● Long time exposure in light of VAHTS SYBR qPCR Master Mix will reduce total fluorescence and may result in ΔCt values >3.6.
(4). Poor reproducibility between replicates.
● Poor accuracy in liquid handling.
● All reagents should be thawed and mix thoroughly before use.
● Ensure that the appropriate ROX reference dye was used.
(5). ΔCt of library dilutions is not within expected range
● Poor accuracy in liquid handling.
● All reagents should be thawed and mix thoroughly before use.
● The library is difficult to amplify, i.e. the library is extremely GC- / AT-rich or has a long average fragment length (>1 kb).
● The library has degraded. Prepare fresh dilutions of library and keep on ice during reaction setup.
(6). Concentrations calculated from different library dilutions differ by more than 10%
● Poor accuracy in liquid handling.
● All reagents should be thawed and mix thoroughly before use.
● The library is difficult to amplify, i.e. the library is extremely GC-/AT-rich or has a long average fragment length (>1 kb).
● The library has degraded. Prepare fresh dilutions of library and keep on ice during reaction setup.
(7). Library dilutions fall outside of effective range of standard curve.
● If Ct (diluted library) < Ct (DNA Standard 1), the library dilution is not sufficient, usually happening in over-amplified libraries. Increase library dilution factor and
repeat quantification reaction.
● If Ct (diluted library) > Ct (DNA Standard 1), the library is over-diluted or the library preparation fails. The Ct value of a conventional library with a dilution factor
around 1:10,000 should not exceed the Ct value of DNA Standard 6. Decrease the library dilution factor and repeat quantification reaction.
(8). The Ct of DNA Standard 1 appears abnormal
● Inappropriate Baseline setting may delay Ct value of DNA Standard 1, further affecting the calculation of PCR efficiency. Manually adjust the baseline to 1-3
cycles.
● Ensure that the appropriate ROX reference dye was used.
(9). Samples of DNA Standards is amplified, but sample of libraries are not. Or significant difference in Ct between DNA Standards and Library Samples.
● The library molecules does not contain the appropriate adapter sequences for the quantification primers to bind to.
● The library the library is over-diluted. Decrease the library dilution factor and repeat quantification reaction.
● The library has degraded. Prepare fresh dilutions of library and keep on ice during reaction setup.
There are no products listed under this category.
